Search for Present/Absent Expression Calls
Search the present and absent expression calls in the Bgee database. Learn how to search for expression calls in your species, genes, tissues, cells, developmental stages, sexes, and expression data types of interest.
Introduction
Bgee "expression call search" allows you to search all present/absent gene expression calls in the Bgee database. The present/absent expression calls are produced using statistical tests specific to each data type, to identify whether the gene expression level is significantly above the background transcriptional and experimental noise. See the main Bgee publications for details. For instance, for RNA-Seq data, the expression level of selected intergenic regions is used to estimate the background noise in each library, leading to obtain one p-value for the significance of expression for each gene in each library. These p-values are then merged and corrected for multiple testing, to provide a definitive FDR-corrected result for each gene in each condition. This result per gene and condition takes into account all produced p-values, from all available data for all requested data types, in this condition and all its children conditions. More specifically for single-cell RNA-Seq data, data are pseudo-bulked per library and cell type to obtain more signal, no statistical present/absent calls are produced per cell, but per cell population (gene count matrices per cell can still be retrieved, in H5AD format, see the experiment search to retrieve such data).
The expression call search can be performed on the Present/Absent expression calls search page. The examples in this tutorial have been generated using Bgee release 15.0. More information about how the present/absent calls are generated can be found on the expression calls documentation page.
Search for present/absent expression calls
The search for expression present/absent calls form allows you to narrow the expression calls displayed to only those of interest.
To start your search, click Show Form. After selecting your species and gene(s) of interest, the full form will become available. You must first select a species to be able to select some genes.
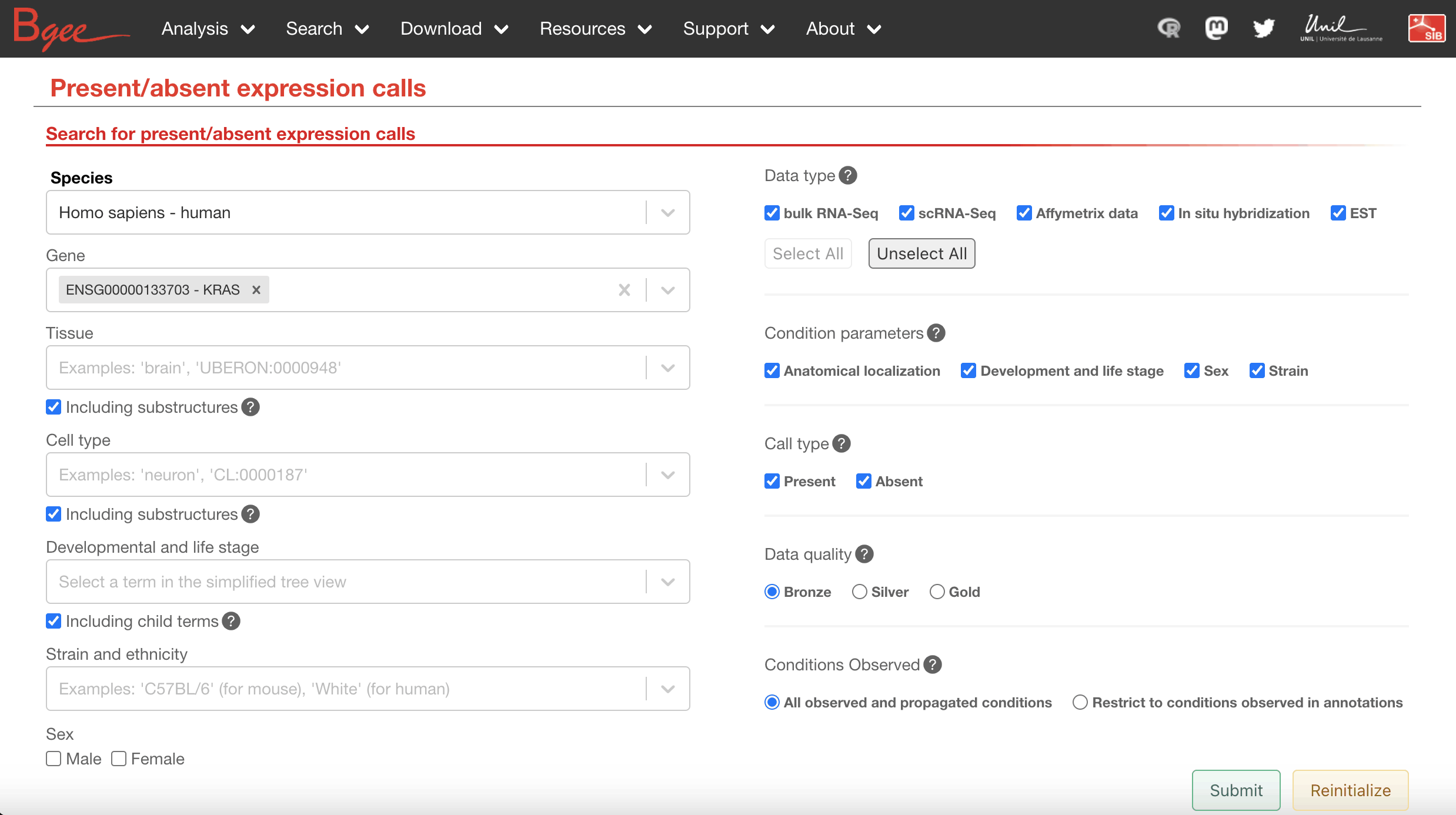
The following filter options are available through the search form:
-
Species (required): Species of interest, either the common name or the Latin name.
-
Gene (required): Gene(s) of interest. You must first select a species for this field to be displayed. You can search using the gene symbol (for instance, HBB) or by the gene identifier (for instance, ENSG00000244734).
-
Tissue: Tissue(s) of interest. You can search using the common name or the ontology ID.
- Checking the including substructures box will include all substructures (child terms) of the tissue(s) you selected. For instance, if you select the term "brain", and this box is checked, all substructures of the brain will also be included in the search.
-
Cell Type: Cell type(s) of interest, either the common name or the ontology ID.
- Checking the including substructures box will include all substructures (child terms) of the cell type(s) you selected.
-
Developmental and Life Stage: Developmental/life stage(s) of interest. Only a simplified tree view of the ontology is displayed. Multiple terms can be selected by reopening the field after clicking on a term.
- Checking the including child terms box will include all child terms of the developmental and life stage(s) you selected.
-
Strain and Ethnicity: Strain(s) of interest, or in humans the ethnicity of interest. Bgee uses the UniProt controlled vocabulary of strains.
-
Sex: Limit sex to male, female, hermaphrodite (depending on the selected species) by checking the corresponding box(es).
-
Data Type: The data type(s) used to produce the expression calls. The calls retrieved will be supported by at least one of the selected data types.
-
Condition Parameter: Select the condition parameters to consider for generating expression calls.
- For example, selecting only "Anatomical localization" will integrate all strain/ethnicity, developmental and life stage, and sex information, into one singular call per gene and anatomical localization.
- Removing a condition parameter, for example "Strain/Ethnicity", will integrate all strain/ethnicity data for a particular anatomical localization, developmental and life stage, and sex, into one singular call per gene.
-
Call type: Limit to presence or absence of expression calls. "Present" means expression level significantly above background transcriptional noise, "absent" means that the level of expression is not significantly different from the background noise.
-
Data Quality: The minimum level of quality in support of the expression call. This level is determined by the FDR-corrected p-values of the statistical test for significant expression of the gene in the reported condition, and in its child conditions.
Results
Filter results
The expression calls can be further filtered by using the fields appearing above the result table after submitting the form. Filters allow you to select actual values present in your results. For instance, if you request in the form to retrieve calls in "brain" including all its substructures, then the filter "Anat. entity and cell type" will display all the brain tissues actually retrieved in the resulting calls.

The following filters can be applied:
- Anat. entity and cell type: Anatomical entity (tissue) or cell type. Selection of multiple terms is supported. You can search using common terms, or the ontology ID.
- Developmental and life stage: You can search using common names, or the ontology ID.
- Sex: select any, female, male, hermaphrodite.
- Strain/Ethnicity: select strain(s)/ethnicity(ies) of interest, including wild type.
Click apply filters to apply the filters you selected. Click the trash icon to remove the selected filters.

Export results
Below the Filter field, the export current page in TSV button allows you to download the results table as a tabulated file. Please note that only the displayed entries will be downloaded in the TSV.
Below the export current page in TSV button, you can change the number of lines visible in the results table with the Show N entries dropdown. The default value is 50 but can be increased up to 1000.
Description of results

-
Gene ID: Gene identifier used to uniquely label each gene.
-
Gene name: It represents the commonly used name for the given gene. For example, in humans the gene names are approved and controlled by the HUGO Gene Nomenclature Committee (HGNC).
-
Present/Absent call: This column shows whether the given gene is considered expressed based on various evidence given by Bgee data types and statistical methods in the specified condition or if the gene was not shown to be active above the background noise.
-
Call quality: Determines how confident Bgee is of the presence/absence call given to the expression state of the gene in that condition. It scale goes from bronze for less confident calls to silver or gold for more strongly supported expression calls.
-
FDR: P-value, corrected for multiple testing by false discovery rate (FDR), of the test of significance used by Bgee to determine the expression state of the gene (various statistical methods are used to determine the p-value based on the data types and for merging results from different data types).
-
Expression score: This value represents the normalized expression level of the gene in that condition after merging all supporting data types. It is scaled from 0, meaning this gene was on average the one with the lowest expression, to 100 which indicates that this gene has high expression in that condition compared to all other genes. Briefly, the expression scores are based on non-parametric statistics, where conditions are ranked based on the gene expression levels. These ranks are then normalized across conditions, genes, and data types, and transformed into an expression score going from 0 (= low expression) to 100 (= high expression) for a more intuitive use. This non-parametric statistics allow comparison of gene expression in a quantitative manner across species and conditions, without requiring batch-correction procedures.
-
Expression score confidence: Bgee's assessment of how confident we are on the validity of the expression score results. There are two possible values, high or low. Our estimation of the validity of the score is determined by looking at the data types used for the expression score calculations. If the score is low and only non-quantitative methods such as in situ hybridization or EST are used, we set its value to low.
-
Supporting data types: In this column, we have 5 squares each representing a different method which Bgee integrates to make the presence/absence calls. The data types that were used to produce the calls of each row is colored in green, all squares in gray show the data types not used for the call either because no data was available for that method in the selected gene and condition, or those data types were filtered out by the user in order to make the expression calls.
-
Anat. entity ID: Controlled vocabulary describing anatomical entity of the samples (UBERON, CL or species specific term) used to make the call. Clicking on the Uberon link brings you to Ontology search page for that Uberon ID where you will be able to see all related substructures or parent terms.
-
Anat. entity name: Common name of the anatomical entity of the sample, for example, "lung".
-
Cell type ID: ID of the cell type of the sample. Clicking the link will give you more information on that cell type by bringing you to the Ontology origin where you can see parent and child terms or cell type function.
-
Cell type name: Common name of the cell type of the sample, such as "embryonic cell".
-
Stage ID: ID of the developmental and life stage of the sample. By clicking on the link you can have more information from the ontology.
-
Stage name: Common name of the developmental and life stage of the sample, such as "adult". You can either have highly precise life stage information such as 4 days old, or if the authors of the libraries used did not have highly precise terms, the most precise parent term is used, in some cases such term can be "life cycle".
-
Sex: Annotation of the sex of the sample (male, female, any, hermaphrodite, mixed).
-
Strain/Ethnicity: Annotation of the specific strain/ethnicity from which the samples were taken, for example C58/J or BXD6.
-
Species: Describes the species name in binomial nomenclature (scientific name).
-
See supporting curated data: Here you can access the datasets and processed expression values used to generate and support the presence/absence calls.